Professional Documents
Culture Documents
Experiment 6
Uploaded by
Shalfiq Mat ZariCopyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Experiment 6
Uploaded by
Shalfiq Mat ZariCopyright:
Available Formats
Lab Manual
CLB 10003 Biology Jan 2013
SECTION OF BIO-ENGINEERING
TECHNOLOGY, UniKL MICET
EXPERIMENT 6: ELECTROPHORESIS AND DNA QUANTIFICATION
Objective: To learn the basic methods used to check the quality of extracted DNA and to
further with DNA quantification
Solutions required
1) 10 X TBE (Tris Borate EDTA). Generally diluted to 1X for use:
2) Sample DNA (from previous lab)
3) 1X TBE agarose gel (1%)
4) Lambda HindIII DNA Marker/ladder
5) Ethidium bromide (EtBr) : HAZARD!!! Wear double gloves when handling.
Experiment Procedures
A.
Electrophoresis of a DNA sample of unknown concentration with a known
standard.
1. Place the gel plate into gel mould, position the comb and ensure that the gel is
horizontal check with a spirit level if necessary.
2. Prepare a 1% agarose gel in 40 ml of 1x TBE. Heat the mixture in a microwave
oven until completely dissolved
3. Pour agarose onto gel tray and allow it to set for least 20 min.
4. Place the gel into the electrophoresis tank. Carefully remove the comb and pour 1x
TBE (same as the buffer that was used to make gel) until the gel is completely
covered.
1
Lab Manual
CLB 10003 Biology Jan 2013
5. Mix 2 l loading dye with 5 l DNA and load it into the well.
6. Mix 1l of Hindlll DNA marker with 1l of loading dye and then load it into one
of the wells.
7. Run the gel at 70 V for one hour or until the dye is about 2/3 from the origin.
8. Stain the gel with ethidium bromide (1 l ethidium bromide in 100 l ddH 2O
(HAZARD!!!!) for 10 minutes.
9. Stain the gel with distilled water for 10 minutes.
10. Illuminate the gel with UV light (CAUTION UV LIGHT IS HAZARDOUS!!!
WEAR MASK OR UV PROTECTION GLASSES IF EXPOSED TO UV
LIGHT).
11. Photograph the gel under the UV.
12. Compare the intensity of the DNA bands of the samples with the intensity of the
DNA Marker bands.
13. Quality of extracted DNA can also be assessed by looking at the gel. Good quality
DNA will show as a sharp intense band. Degraded DNA extracts will show various
degree of smearing. (Refer to figure provided below).
Lane 1
Lane 2
Lane 3
Lane 4
Lane 5
Lane 6
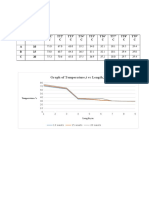
Figure 1 : Agarose gel analysis of genomic DNA isolated from fish tissue.
(From left to right) Lane 1 & 2: Good DNA quality; Lane 3 & 4: DNA with
RNA; Lane 5 & 6: degraded DNA.
Lab Manual
B.
CLB 10003 Biology Jan 2013
Spectrophotometric determination of DNA concentration
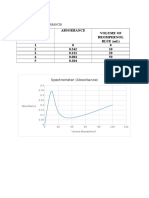
Dilute 3 l of DNA with 150 with deionised water and read at A230, A260 and A280. The
A260/A280 ratio provides an estimate of the purity of the DNA. In a pure sample, this ratio
is approximately 1.8. Lower values indicate protein or phenol contamination. A230 should
be less than A260 and may be the same as A280. High A230 reading indicates that residual
phenol remains in the preparation. An A260 of 1 corresponds to approximately 50 l/ml of
double-stranded DNA in a 1 cm quartz cuvette. Nucleic acid concentration is calculated
as follows:
DNA concentration (ng/l) = A260 X 50 X dilution factor (150 l/3 l)
DNA purity = A260/ A280
Table 1: Common problems in DNA extraction and appropriate solutions.
Discussion
Calculate the DNA concentration and DNA purity.
You might also like
- Agarose Gel ElectrophoresisDocument7 pagesAgarose Gel ElectrophoresisMahathir Mohmed79% (14)
- Agarose Gel ElectrophoresisDocument5 pagesAgarose Gel ElectrophoresisSanmugam SubbaramaniamNo ratings yet
- Agarose Gel Electrophoresis (Full Report)Document10 pagesAgarose Gel Electrophoresis (Full Report)El LisNo ratings yet
- Dna Quality and Quantification: Genetic LabDocument3 pagesDna Quality and Quantification: Genetic LabNgọc Phương Anh NguyễnNo ratings yet
- Basic Principle: To Study The Isolation of Plant Genomic DNA by Using Modified CTAB MethodDocument35 pagesBasic Principle: To Study The Isolation of Plant Genomic DNA by Using Modified CTAB MethodPAWANKUMAR S. K.No ratings yet
- Departments: SDMVM'S College of Agricultural BiotechnologyDocument45 pagesDepartments: SDMVM'S College of Agricultural BiotechnologyPAWANKUMAR S. K.No ratings yet
- DNA Isolation, Restriction, Visualitation, and QuantificationDocument20 pagesDNA Isolation, Restriction, Visualitation, and QuantificationSonianto kuddi100% (5)
- Agarose Gel Electrophoresis PDFDocument3 pagesAgarose Gel Electrophoresis PDF9001 Trisha BakshiBBT2No ratings yet
- Genomic Dna ManualDocument7 pagesGenomic Dna ManualZafran KhanNo ratings yet
- Total DNA Extraction and Quantification: Biomolecular TechniquesDocument20 pagesTotal DNA Extraction and Quantification: Biomolecular TechniquesIzzy JelinNo ratings yet
- DNA Quantification ProtocolDocument4 pagesDNA Quantification Protocolme_dayakar100% (1)
- Agarose Gel Electrophoresis (AGE)Document16 pagesAgarose Gel Electrophoresis (AGE)Nenita AlonzoNo ratings yet
- The Detection of Genes in Fungi by PCR Amplification and Agarose Gel ElectrophoresisDocument12 pagesThe Detection of Genes in Fungi by PCR Amplification and Agarose Gel ElectrophoresisAnsah Samuel SafoNo ratings yet
- 18BTC105J Molecular Biology Laboratory Manual PDFDocument65 pages18BTC105J Molecular Biology Laboratory Manual PDFChris PenielNo ratings yet
- AGAROSE GEL ELECTROPHORESIS - Assignment (AutoRecovered)Document19 pagesAGAROSE GEL ELECTROPHORESIS - Assignment (AutoRecovered)Sathvik BangiramaneNo ratings yet
- Tatie Molecular Genetics Write UpDocument11 pagesTatie Molecular Genetics Write UpYOLANDA NYARADZO MUPITANo ratings yet
- Transformation ConfirmationDocument5 pagesTransformation ConfirmationMuhammad MoeezNo ratings yet
- Gel ElectrophoresisDocument4 pagesGel ElectrophoresisSharmilaJeromeNo ratings yet
- Activity 2. Gel Extraction and PurificationDocument5 pagesActivity 2. Gel Extraction and Purificationgarcia.zaniabNo ratings yet
- DNA Barcoding, Amplification, and Sequencing Lab ReportDocument12 pagesDNA Barcoding, Amplification, and Sequencing Lab ReportRichie JustinNo ratings yet
- Gel ElectrophoresisDocument6 pagesGel ElectrophoresisIshtiaque HossainNo ratings yet
- BCH 212 Last PracDocument10 pagesBCH 212 Last PracNOLUBABALONo ratings yet
- Ceb Practical: 1. TitleDocument8 pagesCeb Practical: 1. TitleKischa DoesesNo ratings yet
- Restriction Enzyme AnalysisDocument8 pagesRestriction Enzyme AnalysisBrenner Celegans0% (1)
- Agarose Gel Electrophoresis Age 1Document3 pagesAgarose Gel Electrophoresis Age 1ShreyaNo ratings yet
- Agarose Gel ElectrophoresisDocument6 pagesAgarose Gel ElectrophoresisAhmad A. ArabiNo ratings yet
- Dna Restriction AnalysisDocument12 pagesDna Restriction Analysisapi-311220353No ratings yet
- Lab Report BET305 - Rahmah Hayati Binti Mohd FauziDocument11 pagesLab Report BET305 - Rahmah Hayati Binti Mohd Fauzirahmah hayatiNo ratings yet
- Plasmid DNA Extraction and Agarose Gel ElectrophoresisDocument5 pagesPlasmid DNA Extraction and Agarose Gel ElectrophoresisPrithvi ThakurNo ratings yet
- DNA Extraction: Qualitative Estimation of Genomic DNADocument32 pagesDNA Extraction: Qualitative Estimation of Genomic DNAPAWANKUMAR S. K.No ratings yet
- Experiment:5: Agarose Gel ElectrophoresisDocument16 pagesExperiment:5: Agarose Gel ElectrophoresisANKIT KUMARNo ratings yet
- IBO 2012 Biology OlympiadDocument49 pagesIBO 2012 Biology OlympiadAbhinav ShuklaNo ratings yet
- Molecular Biology Lab Manual FinalDocument19 pagesMolecular Biology Lab Manual FinalAnupriyaNo ratings yet
- Exercise 7 Agarose Gel Electrophoresis: Cell and Molecular Biology LaboratoryDocument8 pagesExercise 7 Agarose Gel Electrophoresis: Cell and Molecular Biology LaboratoryDham DoñosNo ratings yet
- Agaroe Gel Electrophoresis-2023Document34 pagesAgaroe Gel Electrophoresis-2023abrahamkwandor5678No ratings yet
- CTAB ExtractionDocument3 pagesCTAB ExtractionJanikaa Singaravel MuruganNo ratings yet
- Deoxyribonucleic Acid (DNA), Sodium Salt (D1626) - Product Information SheetDocument2 pagesDeoxyribonucleic Acid (DNA), Sodium Salt (D1626) - Product Information SheetJulioNo ratings yet
- Characterization of Protien and Dna by Gel Electrophoresis Tayyba Tariq 2017-2329Document17 pagesCharacterization of Protien and Dna by Gel Electrophoresis Tayyba Tariq 2017-2329Tayyba TariqNo ratings yet
- Electrophoresis LabDocument3 pagesElectrophoresis LabghustoNo ratings yet
- MAN0013004 GeneRuler 1kb DNALadder 250ug UGDocument3 pagesMAN0013004 GeneRuler 1kb DNALadder 250ug UGBalaji AngamuthuNo ratings yet
- Agarose Gel ElectrophoresisDocument11 pagesAgarose Gel ElectrophoresisAbrar 111No ratings yet
- ProtquantdnaDocument2 pagesProtquantdnaWikan SariNo ratings yet
- DNA Quality-Spectrophotometry and ElectrophoresisDocument5 pagesDNA Quality-Spectrophotometry and Electrophoresislovina candra kirana100% (1)
- Experiment 4Document5 pagesExperiment 4imenmezhoud1122No ratings yet
- Lab 2 ds180 Genotyping LabDocument7 pagesLab 2 ds180 Genotyping Labapi-342081300No ratings yet
- Generuler 1 KB Dna Ladder: Contents and StorageDocument2 pagesGeneruler 1 KB Dna Ladder: Contents and StorageNICOLAS IGNACIO VALENZUELA SAYESNo ratings yet
- DNA Extraction Lab ReportDocument7 pagesDNA Extraction Lab ReportNazurah IbrahimNo ratings yet
- Agarose Gel ElectrophoresisDocument6 pagesAgarose Gel ElectrophoresisJêyà BharathìNo ratings yet
- DNA Lab ReportDocument6 pagesDNA Lab ReportKarina GaidukNo ratings yet
- DNA Abs and FluorescenceDocument6 pagesDNA Abs and FluorescenceDaniela SeczonNo ratings yet
- Agarose Gel Electrophoresis DNADocument5 pagesAgarose Gel Electrophoresis DNAAli Hasan100% (1)
- Module II - HybridizationDocument13 pagesModule II - HybridizationAnanya SinghNo ratings yet
- Molecular Biology - Amity University RajasthanDocument13 pagesMolecular Biology - Amity University Rajasthanabash_u1No ratings yet
- 5. Agarose gel مختبرDocument2 pages5. Agarose gel مختبرAbrar 111No ratings yet
- RNA PolymeraseDocument30 pagesRNA PolymeraseRaghu RokadaNo ratings yet
- Lab 4 Agarose Gel ElectrophoresisDocument6 pagesLab 4 Agarose Gel Electrophoresisjimmy_ncop4702No ratings yet
- Title of ExperimentDocument13 pagesTitle of ExperimentMuskan KhanNo ratings yet
- Title of ExperimentDocument13 pagesTitle of ExperimentHồ Thanh MaiNo ratings yet
- Presented By: Sathvik B S 21L00376 BSC Biotechnology 4 SemesterDocument18 pagesPresented By: Sathvik B S 21L00376 BSC Biotechnology 4 SemesterSathvik BangiramaneNo ratings yet
- Experiment 7Document4 pagesExperiment 7Shalfiq Mat ZariNo ratings yet
- Int - Area Under CurveDocument5 pagesInt - Area Under CurveShalfiq Mat ZariNo ratings yet
- CLB1103 - Biology of Cell: Chapter 1-1Document27 pagesCLB1103 - Biology of Cell: Chapter 1-1Shalfiq Mat ZariNo ratings yet
- Chapter 1 Introduction To Eng CalDocument17 pagesChapter 1 Introduction To Eng CalShalfiq Mat ZariNo ratings yet
- Question Exp3 BiologyDocument1 pageQuestion Exp3 BiologyShalfiq Mat ZariNo ratings yet
- 5.0 Discussion: M V M VDocument2 pages5.0 Discussion: M V M VShalfiq Mat ZariNo ratings yet
- Question Exp3 BiologyDocument1 pageQuestion Exp3 BiologyShalfiq Mat ZariNo ratings yet
- Steam TableDocument2 pagesSteam TableShalfiq Mat ZariNo ratings yet
- Tutorial 2 (2013)Document7 pagesTutorial 2 (2013)Shah AhmadNo ratings yet
- RESULT N Tutorial Heat ConductionDocument6 pagesRESULT N Tutorial Heat ConductionShalfiq Mat ZariNo ratings yet
- 4.0 Result: Spectrometer (Absorbance)Document2 pages4.0 Result: Spectrometer (Absorbance)Shalfiq Mat ZariNo ratings yet
- Report Exp 2 Heat Transfer Discussion (Heat Convection)Document2 pagesReport Exp 2 Heat Transfer Discussion (Heat Convection)Shalfiq Mat ZariNo ratings yet
- 2.0 Performance Evaluation of Mineral OperationsDocument35 pages2.0 Performance Evaluation of Mineral OperationsVitu Verctor ViyuyiNo ratings yet
- Salama YoussefDocument1 pageSalama YoussefYoussef SalamaNo ratings yet
- Basics of Hydrotreating Catalyst Sulfiding - Reactor Resources - Sulfiding Services, Alumina, Metal Reclamation, CatalystsDocument5 pagesBasics of Hydrotreating Catalyst Sulfiding - Reactor Resources - Sulfiding Services, Alumina, Metal Reclamation, Catalystsonizuka-t2263No ratings yet
- K R I T I L E N® Masterbatches: Additives Technical InformationDocument10 pagesK R I T I L E N® Masterbatches: Additives Technical InformationAnas AbdoNo ratings yet
- Sol Gel Synthesis of Nanocrystalline Magnesium Fluoride Its Use in The Preparation of MgF2 Films and MgF2 SiO2 Composites 1996 Chemistry of MaterialsDocument8 pagesSol Gel Synthesis of Nanocrystalline Magnesium Fluoride Its Use in The Preparation of MgF2 Films and MgF2 SiO2 Composites 1996 Chemistry of MaterialskarthikdhadalaNo ratings yet
- Datasheet For Vent ScrubberDocument5 pagesDatasheet For Vent ScrubbercliffrajjoelNo ratings yet
- Rser D 16 02436R1Document88 pagesRser D 16 02436R1Jitender KaushalNo ratings yet
- GENBIO2 - Lesson - The Central Dogma of Molecular BiologyDocument2 pagesGENBIO2 - Lesson - The Central Dogma of Molecular BiologyJazmaine SimbulanNo ratings yet
- Fiitjee PDT Courseware FTRE - 8th Moving To 9th-PHY-ColourDocument10 pagesFiitjee PDT Courseware FTRE - 8th Moving To 9th-PHY-ColourYASHNo ratings yet
- Shell Gadus S3 Wirerope: Performance, Features & Benefits Main ApplicationsDocument2 pagesShell Gadus S3 Wirerope: Performance, Features & Benefits Main ApplicationsptscmscNo ratings yet
- Numerical Analysis For Energy Performance Optimization of Hollow Bricks For Roofing. Case Study - Hot Climate of AlgeriaDocument10 pagesNumerical Analysis For Energy Performance Optimization of Hollow Bricks For Roofing. Case Study - Hot Climate of Algeriamah0809No ratings yet
- T-Technology Pintér WorksDocument5 pagesT-Technology Pintér Worksmig232323No ratings yet
- Assignment 1Document3 pagesAssignment 1Miraj savani100% (1)
- (A6) Durability of Concrete With Different Mineral Admixtures A ReviewDocument12 pages(A6) Durability of Concrete With Different Mineral Admixtures A ReviewAbd El-nour RamdNo ratings yet
- ZL 440 Product Data SheetDocument3 pagesZL 440 Product Data SheetVishal ChudasamaNo ratings yet
- Watford 2001Document7 pagesWatford 2001CARDIO 2019No ratings yet
- DOC316.53.01122 11ed PDFDocument6 pagesDOC316.53.01122 11ed PDFJean Carlos ArangoNo ratings yet
- Space WeatherDocument36 pagesSpace WeatherMat MinNo ratings yet
- Stockmann 2016Document12 pagesStockmann 2016Barbara SilvaNo ratings yet
- An 31.2 Care and Maintenance of Hot Dip GalvanizingDocument4 pagesAn 31.2 Care and Maintenance of Hot Dip GalvanizingnarmathaNo ratings yet
- Gallic AcidDocument28 pagesGallic AcidDolih GozaliNo ratings yet
- Evs MCQDocument8 pagesEvs MCQraj kundraNo ratings yet
- Evamarine: Drying Time Set-To-Touch Hard Dry Painting Interval Min MaxDocument1 pageEvamarine: Drying Time Set-To-Touch Hard Dry Painting Interval Min MaxcelescopitoNo ratings yet
- BHMA Finish ChartDocument5 pagesBHMA Finish ChartRey Eduard Q. UmelNo ratings yet
- Sop of UV HPLCDocument5 pagesSop of UV HPLCSachin S RaneNo ratings yet
- Ecosystem CyclesDocument11 pagesEcosystem CyclesMohammad ShormanNo ratings yet
- 1.7 Evaporative Air Cooling EquipmentDocument8 pages1.7 Evaporative Air Cooling EquipmentRio BananNo ratings yet
- Process Flow Chart Dairy ProductsDocument5 pagesProcess Flow Chart Dairy Productslokesh jainNo ratings yet
- Flash Outokumpu Continuous Converting ProcessDocument15 pagesFlash Outokumpu Continuous Converting ProcessSimón BaezaNo ratings yet
- TDS - Opulyn 301 - Ingles - H&LDocument3 pagesTDS - Opulyn 301 - Ingles - H&LRicardo BohorquezNo ratings yet