Professional Documents
Culture Documents
Purification and Characterization of Two Isoforms From Candida Rugosa Lipase B
Uploaded by
FrontiersOriginal Description:
Original Title
Copyright
Available Formats
Share this document
Did you find this document useful?
Is this content inappropriate?
Report this DocumentCopyright:
Available Formats
Purification and Characterization of Two Isoforms From Candida Rugosa Lipase B
Uploaded by
FrontiersCopyright:
Available Formats
Biotechnology Letters 22: 1291–1294, 2000.
© 2000 Kluwer Academic Publishers. Printed in the Netherlands.
1291
Purification and characterization of two isoforms from Candida rugosa
lipase B
Cristina López, Nelson P. Guerra & M. Luisa Rúa∗
Area de Bioquı́mica e Bioloxı́a Molecular, Facultade de Ciencias de Ourense, Universidade de Vigo,
32004 Ourense, Spain
∗ Author for correspondence (Fax: +34 988 387001; E-mail: mlrua@uvigo.es)
Received 12 May 2000; Revisions requested 23 May 2000; Revisions received 20 June 2000; Accepted 22 June 2000
Key words: Candida rugosa, characterization, isoforms, lipase B, purification
Abstract
Two isoforms of Candida rugosa lipase B (LB1 and LB2) were purified by anionic exchange chromatography. The
lipases had the same N-terminal sequence, carbohydrate content and pH and thermal stability but different pIs and
significant differences in their activities against different p-nitrophenol esters and triacylglycerides.
Introduction Materials and methods
Candida rugosa secretes a mixture of lipase isoen- Materials
zymes which have been extensively used in biotrans-
formations. Lotti & Alberghina (1996) have elucidated Lipase type VII from C. rugosa, endoproteinase Glu-
seven lipase gene sequences of C. rugosa (LIP1- C and triolein emulsion and p-nitrophenyl esters used
LIP7), five of them (LIP1-LIP5) having been fully as substrates were from Sigma. Tributyrin and tri-
characterized. Comparison of the predicted amino acid acetin were from Fluka (Deisenhofen, Germany). N-
sequences reveals a close similarity among the five glycosidase F deglycosylation kit from Boehringer
lipases, ranging between 77% and 88% identity for Mannhein (Mannhein, Germany). Ion-exchange col-
pairs of proteins. However, an important variability umn (Mono P HR 5/5), desalting columns (PD-10)
in several positions around the active site that could and gels for isoelectric focusing were from Pharmacia
influence interactions with the substrate have been re- (Sweden). All other chemicals used were of analytical
ported (Lotti & Alberghina 1996). Only three lipase grade.
isoenzymes have been identified in C. rugosa lipase
(Sigma type VII), the commercial extract most fre- Purification
quently used in biotransformations. Diczfalusy et al. Purification of lipase B was carried out as described by
(1997) detected Lip2 and two closely related enzymes: Rúa & Ballesteros (1994). Concentrated lipase B was
CEL1 and CEL3 both being the product of LIP1 gene. loaded on a Mono P HR5/5 column equilibrated with
Aditionally, Rúa & Ballesteros (1994), using a sim- 25 mM histidine/HCl buffer (pH 6.1). The column was
pler method, now used in several laboratories, purified eluted applying a linear gradient of 0.150–0.275 M
Lip3 and a mixture of 4 isoforms with different pIs NaCl in 20 ml of equilibration buffer. The obtained
named as lipase B, which is the main fraction in Sigma lipases were desalted using PD-10 columns equili-
extracts (70%) (Rúa et al. 1993). Nevertheless, it brated in 25 mM Tris/HCl (pH 7.5) following the
is not known whether or not the isoforms are true manufacturer’s instructions.
isoenzymes. The aim of this work was the separa-
tion of lipase B isoforms in order to determine their
differences at molecular and activity level.
1292
Fig. 1. Chromatography on Mono P HR5/5. 500 µl lipase B was loaded onto the column equilibrated in 25 mM histidine/HCl buffer (pH 6.1).
Bound lipases were eluted with a linear gradient of NaCl (0.150–0.275 M) in 20 ml of equilibration buffer. Flow rate: 1 ml min−1 ; fractions:
0.5 ml.
Enzyme assays by SDS-PAGE in 10% polyacrylamide gels, on a ver-
Lipase activity was assayed in a Metrohm pH-stat at tical slab mini gel apparatus (Model SE 250; Pharma-
30 ◦ C in 5 mM Tris/HCl (pH 7.0) containing 0.1 M cia Biotech) and stained for proteins with Coomassie
CaCl2 , using 1 M triacetin (TA), 0.11 M tributyrin Brillant Blue R-250.
(TB) and 0.5 ml commercial emulsion of triolein (TO).
The final volume of reaction mixture was 15 ml. The Isoelectric focusing (IEF)
esterase activity was followed spectrophotometrically Commercial gels in the pH-range 3.5–9.5 were em-
at 348 nm using p-nitrophenol esters as substrates. The ployed. The electrophoresis was performed on a Mul-
assay was performed at 30 ◦ C with 1 mM of each p- tiphor Electrophoresis System (Pharmacia Biotech)
nitrophenol ester in 50 mM sodium phosphate buffer following the manufacturer’s intructions and stained
(pH 7.0) containing 0.3% (w/v) Triton X-100 and 4% with Coomassie Brillant Blue R-250.
(v/v) acetone. One activity unit was defined as the
quantity of enzyme necessary to release 1 µmol prod- Peptide mapping
uct per min. To compare statistically the activity mean Proteolytic digestion of denatured lipase samples was
values of LB1 and LB2 against different substrates, a performed as described by Cleveland et al. (1977).
hypothesis test of one tail was used. Denatured lipase samples, dissolved in 50 mM am-
monium bicarbonate (pH 7.8) containing 0.2% (w/v)
Protein concentration SDS were incubated for 1 h at 37 ◦ C with endopro-
Protein concentration was determinated using the teinase Glu-C, keeping the substrate to enzyme ratio
Lowry method with bovine serum albumin as stan- of 50. The reaction was stopped adding one volume
dard. of electrophoresis sample buffer and boiling at 95 ◦ C
for 3 min. About 20 µl (20 µg) of each sample were
Amino terminal sequence analysis analysed by SDS-PAGE.
The purified proteins were sequenced directly from
solutions in a Beckman LF3000 sequencer.
Enzymatic deglycosylation of the lipases
Digestion of lipase samples was carried out using the
N-glycosidase F deglycosylation kit, according to the
manufacturer’s instructions. Samples were analysed
1293
Table 1. Summary of the purification.
Fraction Total protein Specific activity Purification Yield
(mg) (U mg−1 ) (fold) (%)
Lipase B 2.7 1080 1 100
Mono P HR5/5
LB1 1.0 700 0.7 28
LB2 0.5 385 0.4 7
Comparison of lipase B isoforms at the molecular
level
Lipases LB1 and LB2 showed different pIs: 4.8 (LB1)
and 4.7 (LB2) but same carbohydrate content (2.5%).
The N-terminal sequences were identical up to the
20th amino acid (APTATLLANGDTITGLNAII). Ac-
cording to the published sequences, lipases LB1 and
LB2 could be the product of the genes LIP1, LIP4 and
LIP5 but not LIP3 (lysine in position 5) or LIP2 (va-
line in position 19). Isoenzymes LB1 and LB2 were
digested with endoproteinase Glu-C but the resulting
peptides were identical for both enzymes (not shown).
Effect of pH and temperature on the lipase activity
and stability
The effect of pH and temperature on the activity and
stability of LB1 and LB2 was studied using triolein
Fig. 2. Isoelectric focusing. Lipase B (lane 1); lipase LB1 (lane as substrate. No significative differences could be ob-
2) and lipase LB2 (lane 3). Gels were stained for protein with
Coomassie Blue R-250. served between the purified lipases: the optimum pH
and temperature were 7.0 and 30 ◦ C, respectively, for
both enzymes. Both showed high stability at pH 5.0
Results and 7.0 whereas at pH 9.0 were completely inacti-
vated after 30 min incubation.Their thermal stabilities
Purification of lipase B isoforms at 40 ◦ C were almost identical.
Lipase B from C. rugosa was purified as described by
Activity on triacylglycerides and p-nitrophenyl esters
Rúa & Ballesteros (1994). Only the two major pro-
tein bands detected by the authors could be observed LB1 and LB2 activity increased with the chain-length
by IEF. These proteins were separated on a Mono P of the assayed triacylglycerides (TO>TB>TA) be-
HR5/5 (Figure 1) as described in Materials and meth- ing isoform LB1 significantly more active than LB2
ods. Isoform LB1 (fractions 31–36) was the major one (Table 2). On p-nitrophenyl esters, the catalytic dif-
with a 28% yield being also more active than LB2 ferences between the isoforms were more noticeable
(fractions 38–41) on tributyrin (700 vs 385 U mg−1 ) than with triacylglycerides. Thus, with the exception
(Table 1). The purity of both fractions was checked by of pNPC6, LB2 was significantly more active than
IEF (Figure 2). LB1, being the differences more marked with pNPC4
and pNFC10, which were the best substrates for both
isoforms. The activity profiles passed through two
minima with pNPC3 and pNPC6 (Table 2).
1294
Table 2. Specific activity of lipases LB1 and LB2 on triacylglycerides and various
p-nitrophenyl esters and significance.
Substrate Specific activity (U mg−1 ) te t(α ≤ 0.05) ν
LB1 LB2
Triolein 2040 1860 4.5 2.8 4
Tributyrin 833 725 8.3 4.3 2
Triacetin 29 26 4.5 2.8 4
pNPC3 12 13 3.8 2.2 10
pNPC4 27 49 4.9 2.2 10
pNPC6 15 15 0.0 2.2 10
pNPC10 28 44 3.5 2.2 10
te: experimental Student t; ν: degree freedom; pNPC3, pNPC4, pNPC6 and
pNPC10: p-nitrophenyl propionate, butyrate, caproate and caprate, respectively.
Discussion of minor posttranslational modifications of LIP1 or
the product of different genes. Alberghina & Lotti
In this work a method was developed for the isola- (1997)have reported the existence of at least two addi-
tion of two lipases from the pool of lipase B isoforms tional genes (LIP6 and LIP7) and perhaps up to three
(LB1 and LB2), using anionic exchange chromatogra- more, in the lipase gene family. Thus, taking into ac-
phy. Although some characteristics of LB1 (the major count the significant kinetic differences found between
isoform) and LB2 were similar (N-terminal sequence, LB1 and LB2, it could be suggested that LB2 would
molecular weight, carbohydrate content, peptide frag- be the product of one of these genes. It should be em-
ments and stability) differences in pI and activity on phasized that these two proteins (LB1 and LB2) have
triacylglycerides and p-nitrophenyl esters were found. been identified in all Sigma lots used for more than 8
Thus, LB1 showed significant higher activity on tri- years.
acylglycerides than LB2, whereas LB2 was more
effective to hydrolyze the p-nitrophenyl esters (with
exception of pNPC6 to whom both had identical ac- References
tivity). Our results strongly resemble those reported
by Diczfalusy et al. (1997) for isoforms CEL1 and Alberghina L, Lotti M (1997) Cloning, sequencing and expression
of C. rugosa lipases. In: Rubin B, Dennis E, eds. Methods in
CEL3 although the authors do not provide their pIs Enzymology, Vol. 284. San Diego: Academic Press, pp. 246–260.
thus making difficult a direct comparison with LB1 Cleveland DW, Fischer SG, Kirschner MW, Laemmli U (1977)
and LB2. CEL1 exhibed a markedly higher activity Peptide mapping by limited proteolysis in sodium dodecyl sul-
than CEL3 on triolein and tributyrin, similarly to LB1 fate and analysis by gel electrophoresis. J. Biol. Chem. 252:
1102–1106.
purified in this work. Also, both had the same activ- Diczfalusy MA, Hellman U, Alexson SEH (1997) Isolation of
ity on pNPC6 as isoforms LB1 and LB2. However, carboxylester lipase (CEL) isoenzymes from C. rugosa and iden-
although CEL1 was more active than CEL3 on those tification of the corresponding genes. Arch. Biochem. Biophys.
esters with the shortest acyl chain (C < 6) the opposite 348: 1–8.
Grochulski P, Schrag JD, Douthillier F, Smith P, Harrison D, Rubin
was true for the esters with the longhest acyl chains (C B, Cygler M (1993) Insights into interfacial activation from an
> 6). Apart from this difference, LB1 might be the open structure of C. rugosa lipase. J. Biol. Chem. 268: 12843–
same protein as CEL1 also the major protein in ex- 12847.
Lotti M, Alberghina L (1996) Candida rugosa lipase isoenzymes.
tracts purified by Diczfalusy et al. (1997). As LIP1 has
In: Malcata FX, ed. Engineering of/with Lipases, Dordrecht:
been cristallizated from Sigma extracts and reported to Kluwer Academic Publishers, pp. 115–124.
be the major isoenzyme in this preparation (Grochul- Rúa ML, Ballesteros A (1994) Rapid purification of two lipases
ski et al. 1993) it seems plausible that LB1 and CEL1 from C. rugosa. Biotechnol. Tech. 8: 21–26.
Rúa ML, Díaz-Mauriño T, Fernández VM, Otero C, Ballesteros A
correspond to LIP1. The origin of LB2 remains uncer-
(1993) Purification and characterization of two distinct lipases
tain: as suggested by Diczfalusy et al. (1997) a result from C. cylindracea. Biochim. Biophys. Acta 1156: 181–189.
You might also like
- Novozym 435 Displays Very Different Selectivity Compared To Lipase From Candida Antarctica B Adsorbed On Other Hydrophobic SupportsDocument6 pagesNovozym 435 Displays Very Different Selectivity Compared To Lipase From Candida Antarctica B Adsorbed On Other Hydrophobic SupportsghcoutoNo ratings yet
- Purification and Characterization of A Novel Solvent-Tolerant Lipase FromDocument4 pagesPurification and Characterization of A Novel Solvent-Tolerant Lipase Fromسید حسین عارفیNo ratings yet
- Cloning, Purification and Comparative Characterization of Two Digestive Lysozymes From Musca Domestica LarvaeDocument9 pagesCloning, Purification and Comparative Characterization of Two Digestive Lysozymes From Musca Domestica LarvaebobyjuniorNo ratings yet
- 1990 Feeney Et Al Biochemical and Biophysical Research Communications 30 A Single Amino Acid Substitution in Lactate Dehydrogenase Improves TheDocument6 pages1990 Feeney Et Al Biochemical and Biophysical Research Communications 30 A Single Amino Acid Substitution in Lactate Dehydrogenase Improves Thel4vfeaokf5No ratings yet
- Iai00165 0382Document9 pagesIai00165 0382pabimbinyospaNo ratings yet
- Blue Native Electrophoresis Study On LipasesDocument2 pagesBlue Native Electrophoresis Study On LipasesRubina NeloferNo ratings yet
- Interaction Between Cibacron Blue F3GA and Luteinizing Hormone: A Chromatographic InvestigationDocument15 pagesInteraction Between Cibacron Blue F3GA and Luteinizing Hormone: A Chromatographic InvestigationIOSR Journal of PharmacyNo ratings yet
- Effect of Alpha Lipoic Acid On Leukotriene A4 HydrolaseDocument7 pagesEffect of Alpha Lipoic Acid On Leukotriene A4 HydrolaseChasconaNo ratings yet
- Pseudocholinesterase BoniDocument6 pagesPseudocholinesterase BonibiochemiNo ratings yet
- LFABP and HFABPDocument6 pagesLFABP and HFABPMichiko MatsuoNo ratings yet
- Pnas 84 16 5918Document5 pagesPnas 84 16 5918Ingrid Carolina Salazar AmorósNo ratings yet
- pHInduced Conformational Isomerization of LeghemoglobinDocument7 pagespHInduced Conformational Isomerization of LeghemoglobinPijush BasakNo ratings yet
- Isolation and Characterization of Three Acid Phosphatase Isoenzymes from Wheat GermDocument6 pagesIsolation and Characterization of Three Acid Phosphatase Isoenzymes from Wheat GermBarry WhiteNo ratings yet
- The Effect of Polysorbate 20 and 80 On The Structure and Immunogenicity in Wildtype and Transgenic Mice of Recombinant Human Interferon Alpha2bDocument20 pagesThe Effect of Polysorbate 20 and 80 On The Structure and Immunogenicity in Wildtype and Transgenic Mice of Recombinant Human Interferon Alpha2bMAHESHNo ratings yet
- TMP 1 F28Document6 pagesTMP 1 F28FrontiersNo ratings yet
- 1989 - Tsai Et Al. - Leukotriene Release Enhancing Factor. Purification, Specific Allergen Induction, and Further Biologic PropertiesDocument8 pages1989 - Tsai Et Al. - Leukotriene Release Enhancing Factor. Purification, Specific Allergen Induction, and Further Biologic Propertiespond_1993No ratings yet
- EMB - Carboxypeptidase YDocument2 pagesEMB - Carboxypeptidase YJim Well MartinNo ratings yet
- A Simple HPLC Method For The Simultaneous Analysis of Phosphatidylcholine and Its Partial Hydrolysis Products 1-And 2-Acyl LysophosphatidylcholineDocument5 pagesA Simple HPLC Method For The Simultaneous Analysis of Phosphatidylcholine and Its Partial Hydrolysis Products 1-And 2-Acyl LysophosphatidylcholineChang Woo JongNo ratings yet
- Biochemical Properties of A B-Xylosidase From: Clostridium CellulolyticumDocument4 pagesBiochemical Properties of A B-Xylosidase From: Clostridium CellulolyticumPedro HamannNo ratings yet
- PosterSSCHE2014 ReactEngineeringnCatalysisPosterSessionDocument1 pagePosterSSCHE2014 ReactEngineeringnCatalysisPosterSessionDiana Noor IsmailNo ratings yet
- Production of Extracellular Lipase by The Phytopathogenic Fungus Fusarium Solani Fs1Document6 pagesProduction of Extracellular Lipase by The Phytopathogenic Fungus Fusarium Solani Fs1Xenia MenaNo ratings yet
- Serine Subtilis Strain Marburg: Intracellular Proteinase of Bacillus 168Document7 pagesSerine Subtilis Strain Marburg: Intracellular Proteinase of Bacillus 168Cristian Sneider Pinzon TopalNo ratings yet
- Características Del Aceite Extraído Con Fosfatasa Recombinante LIS KATERINEDocument6 pagesCaracterísticas Del Aceite Extraído Con Fosfatasa Recombinante LIS KATERINEYury MartinezNo ratings yet
- J. Biol. Chem.-1968-Barel-1344-8Document5 pagesJ. Biol. Chem.-1968-Barel-1344-8Nguyễn Ngô SangNo ratings yet
- TMP CA0 DDocument6 pagesTMP CA0 DFrontiersNo ratings yet
- Dykes and Kay 1976Document4 pagesDykes and Kay 1976harisankarhsNo ratings yet
- Short Synthetic Peptides as Inhibitors of Angiotensin Converting EnzymeDocument2 pagesShort Synthetic Peptides as Inhibitors of Angiotensin Converting EnzymeDani VankovaNo ratings yet
- 0065 2571 (72) 90019 2Document20 pages0065 2571 (72) 90019 2Muhammad Akbar SusenoNo ratings yet
- Purification and Characterization of Cold Active Lipase From Psychrotrophic Aeromonas Sp. LPB 4Document6 pagesPurification and Characterization of Cold Active Lipase From Psychrotrophic Aeromonas Sp. LPB 4Carlos Alain Floriano GrandezNo ratings yet
- Biotek 2Document9 pagesBiotek 2cupcupmuahNo ratings yet
- tmp5 TMPDocument8 pagestmp5 TMPFrontiersNo ratings yet
- Intramolecular Coupling of Active Sites in the Pyruvate Dehydrogenase ComplexDocument6 pagesIntramolecular Coupling of Active Sites in the Pyruvate Dehydrogenase Complexanshusharma5No ratings yet
- Silva, E. Ugarte, R. Andrarde, A. Edwards, A. M. J. Photochem. Photobiol., B 1994, 23, 43.Document6 pagesSilva, E. Ugarte, R. Andrarde, A. Edwards, A. M. J. Photochem. Photobiol., B 1994, 23, 43.Hylze ChavesNo ratings yet
- Determination of Enzymatic Activities of PDFDocument5 pagesDetermination of Enzymatic Activities of PDFcarolasbdNo ratings yet
- At Pa Sal BrevisDocument5 pagesAt Pa Sal BrevisNils Huaman CastillaNo ratings yet
- CHEM 151 (Chapter 3)Document4 pagesCHEM 151 (Chapter 3)Chantel AceveroNo ratings yet
- Artigo ImobilizaçãoDocument6 pagesArtigo ImobilizaçãocterrasanNo ratings yet
- Synthesis Soybean: Heme in Root NodulesDocument4 pagesSynthesis Soybean: Heme in Root NodulesEmmilyNo ratings yet
- Synthesis and Characterization of New Amino Acid-Schiff Bases and Studies Their Effects On The Activity of ACP, PAP and NPA Enzymes (In Vitro)Document9 pagesSynthesis and Characterization of New Amino Acid-Schiff Bases and Studies Their Effects On The Activity of ACP, PAP and NPA Enzymes (In Vitro)Pavan SrivastavaNo ratings yet
- Formal Report QualiDocument7 pagesFormal Report QualiAyla DizonNo ratings yet
- Kinetic Resolution of (R, S) - 1,2-O-Isopropylideneglycerol by Esterification With Dry Mycelia of MouldsDocument4 pagesKinetic Resolution of (R, S) - 1,2-O-Isopropylideneglycerol by Esterification With Dry Mycelia of MouldsMathias PradoNo ratings yet
- Phospho-opsin Phosphatase Insight Into Protamine StimulationDocument12 pagesPhospho-opsin Phosphatase Insight Into Protamine StimulationnaNo ratings yet
- The Mechanism Activator Inhibitor: of The Reaction 1 and Tissue PlasminogenDocument5 pagesThe Mechanism Activator Inhibitor: of The Reaction 1 and Tissue PlasminogenReju RoseNo ratings yet
- Stabilization of Serum Albumin by Anti-Inflammatory DrugsDocument8 pagesStabilization of Serum Albumin by Anti-Inflammatory DrugsJariyah AmiliaNo ratings yet
- Fosfatase Soluvel e LeismaniaDocument8 pagesFosfatase Soluvel e LeismaniacoringacravoNo ratings yet
- Chemical Modification of A Cellulase From Aspergillus Niger: Components SystemsDocument8 pagesChemical Modification of A Cellulase From Aspergillus Niger: Components SystemsAprilia Isma DenilaNo ratings yet
- 1 PBDocument5 pages1 PBCamilo Ernesto Araujo BarabasNo ratings yet
- Leucine SolubilityDocument7 pagesLeucine SolubilityClarence AG YueNo ratings yet
- Colour Reaction of Amino AcidsDocument27 pagesColour Reaction of Amino AcidsNicola Faye BronNo ratings yet
- Tea Leaf Polyphenol OxidaseDocument10 pagesTea Leaf Polyphenol OxidaseHồng TrinhNo ratings yet
- Laboratoire de G&Ctique Molhdaire, Unitp de Gtin&Tique, Universiti de Croix Du Sud 5 (Bte 6), B-1348 Louvain-La Neuve, Belj$UmDocument4 pagesLaboratoire de G&Ctique Molhdaire, Unitp de Gtin&Tique, Universiti de Croix Du Sud 5 (Bte 6), B-1348 Louvain-La Neuve, Belj$UmSukanthan RNo ratings yet
- Jamur Tiram 1Document6 pagesJamur Tiram 1Muhammad AkbarNo ratings yet
- Cardiotoxin of The Indian Cobra (Naja Naja) Is A PyrophosphataseDocument7 pagesCardiotoxin of The Indian Cobra (Naja Naja) Is A PyrophosphataseRahul DattaNo ratings yet
- Pmecx 16 - Marcado TotalDocument6 pagesPmecx 16 - Marcado TotalVictor RoticivNo ratings yet
- 1998 Heinfling AEM 64 2788Document6 pages1998 Heinfling AEM 64 2788Erika Canales IdeNo ratings yet
- Brandsma (2008) L Lactis Aminotransferases & DehydrogenasesDocument6 pagesBrandsma (2008) L Lactis Aminotransferases & DehydrogenasesNicolas FlahautNo ratings yet
- Physicochemical properties of amino acidsDocument3 pagesPhysicochemical properties of amino acidsyapyapvinxNo ratings yet
- Arber and LinnDocument7 pagesArber and Linnshubham20No ratings yet
- Lichenysin: I G, J W, R M - D, F PDocument12 pagesLichenysin: I G, J W, R M - D, F PAmanda AlencarNo ratings yet
- Co- and Post-Translational Modifications of Therapeutic Antibodies and ProteinsFrom EverandCo- and Post-Translational Modifications of Therapeutic Antibodies and ProteinsNo ratings yet
- tmpFFE0 TMPDocument6 pagestmpFFE0 TMPFrontiersNo ratings yet
- tmpCE8C TMPDocument19 pagestmpCE8C TMPFrontiersNo ratings yet
- tmp80F6 TMPDocument24 pagestmp80F6 TMPFrontiersNo ratings yet
- tmp3CAB TMPDocument16 pagestmp3CAB TMPFrontiersNo ratings yet
- tmpEFCC TMPDocument6 pagestmpEFCC TMPFrontiersNo ratings yet
- Tmpa077 TMPDocument15 pagesTmpa077 TMPFrontiersNo ratings yet
- Tmp1a96 TMPDocument80 pagesTmp1a96 TMPFrontiersNo ratings yet
- tmp6F0E TMPDocument12 pagestmp6F0E TMPFrontiersNo ratings yet
- tmpF178 TMPDocument15 pagestmpF178 TMPFrontiersNo ratings yet
- tmpC0A TMPDocument9 pagestmpC0A TMPFrontiersNo ratings yet
- tmpF3B5 TMPDocument15 pagestmpF3B5 TMPFrontiersNo ratings yet
- tmpE7E9 TMPDocument14 pagestmpE7E9 TMPFrontiersNo ratings yet
- tmpF407 TMPDocument17 pagestmpF407 TMPFrontiersNo ratings yet
- tmpE3C0 TMPDocument17 pagestmpE3C0 TMPFrontiersNo ratings yet
- tmp6382 TMPDocument8 pagestmp6382 TMPFrontiersNo ratings yet
- tmp72FE TMPDocument8 pagestmp72FE TMPFrontiersNo ratings yet
- tmp60EF TMPDocument20 pagestmp60EF TMPFrontiersNo ratings yet
- Tmp75a7 TMPDocument8 pagesTmp75a7 TMPFrontiersNo ratings yet
- tmp8B94 TMPDocument9 pagestmp8B94 TMPFrontiersNo ratings yet
- tmp37B8 TMPDocument9 pagestmp37B8 TMPFrontiersNo ratings yet
- tmp4B57 TMPDocument9 pagestmp4B57 TMPFrontiersNo ratings yet
- tmp998 TMPDocument9 pagestmp998 TMPFrontiersNo ratings yet
- tmp9D75 TMPDocument9 pagestmp9D75 TMPFrontiersNo ratings yet
- tmpD1FE TMPDocument6 pagestmpD1FE TMPFrontiersNo ratings yet
- tmpB1BE TMPDocument9 pagestmpB1BE TMPFrontiersNo ratings yet
- tmpC30A TMPDocument10 pagestmpC30A TMPFrontiersNo ratings yet
- tmpA0D TMPDocument9 pagestmpA0D TMPFrontiersNo ratings yet
- tmp27C1 TMPDocument5 pagestmp27C1 TMPFrontiersNo ratings yet
- tmp3656 TMPDocument14 pagestmp3656 TMPFrontiersNo ratings yet
- tmp2F3F TMPDocument10 pagestmp2F3F TMPFrontiersNo ratings yet
- ANTHE 2021 (Engineering) Sample PaperDocument17 pagesANTHE 2021 (Engineering) Sample PaperDida CowernNo ratings yet
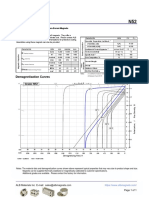
- N52 Grade Neodymium Magnets DataDocument1 pageN52 Grade Neodymium Magnets DataSteve HsuNo ratings yet
- Welding Defects and PreventionDocument2 pagesWelding Defects and PreventionVicky SinghNo ratings yet
- Colder Products Company Full CatalogDocument166 pagesColder Products Company Full CatalogDENNIZNo ratings yet
- Vragen Fasediagrammen Zuivere Componenten PDFDocument3 pagesVragen Fasediagrammen Zuivere Componenten PDFbilberNo ratings yet
- FINAL-PROPOSAL Need Revisions Fire RatingDocument30 pagesFINAL-PROPOSAL Need Revisions Fire RatingMeister MJNo ratings yet
- Introduction To Laser TechnologyDocument31 pagesIntroduction To Laser TechnologyJames Sullivan100% (1)
- Qw-482 Suggested Format For Welding Procedure Specifications (WPS)Document2 pagesQw-482 Suggested Format For Welding Procedure Specifications (WPS)Ravelo Jeisson100% (2)
- Chemistry 2pointsDocument4 pagesChemistry 2pointsjovanniNo ratings yet
- Yemen LNG Upstream Facilities Project: Phase 1 Process Design Basis ManualDocument23 pagesYemen LNG Upstream Facilities Project: Phase 1 Process Design Basis Manualabdoamer.2553No ratings yet
- TOC Application HandbookDocument79 pagesTOC Application Handbookmregalopez3647100% (1)
- Vortex Quantum SeriesDocument34 pagesVortex Quantum SeriesmiguelcNo ratings yet
- Onion Cell Structure Under MicroscopeDocument2 pagesOnion Cell Structure Under MicroscopeAnirudh100% (1)
- Wall Colmonoy Surfacing Alloys Selector ChartDocument3 pagesWall Colmonoy Surfacing Alloys Selector ChartAjimKe'enNo ratings yet
- Lab 2 - StoichiometryDocument4 pagesLab 2 - Stoichiometryapi-272470922100% (3)
- Smelt Water ExplosionsDocument19 pagesSmelt Water Explosionsnmehta67100% (1)
- 맥머리유기화학8판Document1,177 pages맥머리유기화학8판이경식No ratings yet
- ROCKWOOL© Technical InsulationDocument36 pagesROCKWOOL© Technical InsulationHaytham ElsayedNo ratings yet
- Nuclear Power: Pros, Cons and FutureDocument4 pagesNuclear Power: Pros, Cons and FutureSamarthNo ratings yet
- Summary KH2134 Fluid MechanicsDocument4 pagesSummary KH2134 Fluid MechanicsAzman SamerNo ratings yet
- What's New - PV Elite 2018Document28 pagesWhat's New - PV Elite 2018SathiyaseelanNo ratings yet
- IEEE Xplore - SearchResultDocument4 pagesIEEE Xplore - SearchResultSalman KhanNo ratings yet
- Colorimeter Principle PDFDocument2 pagesColorimeter Principle PDFNicholasNo ratings yet
- Disinfection For PH 2Document73 pagesDisinfection For PH 2Boas WayneNo ratings yet
- Entner Duodroff PathwayDocument2 pagesEntner Duodroff PathwayDr. SHIVA AITHALNo ratings yet
- Exp 2 Protein DeterminationDocument5 pagesExp 2 Protein DeterminationNur Fadhilah100% (1)
- Determination of Zinc (Experiment)Document3 pagesDetermination of Zinc (Experiment)Hassan Haider100% (4)
- AG SR SecondaryDocument33 pagesAG SR SecondaryDeepikaNo ratings yet
- Micro 2000 - Deox 2000Document139 pagesMicro 2000 - Deox 2000Achr FFNo ratings yet
- Course Outline For 125:355, Physiological Systems For Biomedical EngineersDocument2 pagesCourse Outline For 125:355, Physiological Systems For Biomedical EngineersbillNo ratings yet